RESEARCH INTERESTS
Signaling by H2S
Hydrogen sulfide (H2S) is one of the simplest sulfur-containing molecules found in cells and since the first report of its potential physiological role, there has been a burgeoning literature on the subject of H2S signaling. Several physiological functions are reported to be regulated by H2S, some of which are vasodilation, neurotransmission, and inflammation (Filipovic et al, Chem Rev, 2018).
H2S is produced by the action of at least three enzymes, cystathionine beta synthase (CBS), cystathionine gamma lyase (CSE) and mercaptopyruvate sulfur-transferase (MST). Differently expressed in different tissues (and cellular compartments) these enzymes control H2S production with different efficiency (Filipovic, Handbook Exp Pharmacol, 2015; Filipovic et al, Chem Rev, 2018).
Signaling by H2S is a new field of research and the exact mechanisms of H2S actions are still under active investigation. My group has been working on elucidating those mechanisms for couple of years. Biologically relevant reactions of H2S could be grouped in three signaling axes: (i) interaction/cross-talk with reactive oxygen (ROS) and nitrogen species (RNS), (ii) interaction with metalloproteins and (iii) oxidative posttranslational modification of cysteine residues (persulfidation or S-sulfhydration) .

Biologically relevant reactions of H2S.
CROSS-TALK WITH NO
Our studies of the interaction of H2S with reactive oxygen and nitrogen species which led to identification of some small inorganic species that could have biological function of their own (Filipovic et al, Biochem J, 2012; Wedmann et al, Inorg Chem, 2015, Ivanovic-Burmazovic & Filipovic, Inorg Chem, 2018). Cross-talk of H2S with S-nitrosothiols led to characterization of the smallest S-nitrosothiol, HSNO, which could easily cross biological membranes, linking S-nitrosation with protein persulfidation (Filipovic et al, JACS, 2012; Miljkovic et al, Angew Chem Int Ed, 2013). The cross-talk of H2S with NO proved to be even more complex (Filipovic et al, J Med Chem, 2013), resulting in formation of NO sibling, HNO (Eberhardt et al, Nat Comm, 2014). We demonstrated that HNO is a powerful stimulator of TRPA1 channels, which results in a release of calcitonin-gene related peptide and subsequent control of blood flow, particularly in brain (Eberhardt et al, Nat Comm, 2014). We used this knowledge to patent a new way for HNO generation that could be used during the acute hypertensive crisis or/and for the treatment of patients suffering from heart failure (WO 2014/122074).
INTERACTION WITH METAL CENTERS
In collaboration with Ruma Banerjee’s team (University of Michigan) we also studied the interaction of H2S with different metalloproteins (Bostelaar et al, JACS, 2016; Ruetz et al, JBC, 2017). Most recently, we observed that in mitochondria, and under the conditions of low oxygen, cytochrome c takes over H2S oxidation, catalyzing persulfidation of surrounding proteins. Cytochrome c catalyzed persulfidation of procaspase 9 resulted in inhibition of apoptosis in cells (Vitvitsky et al, ACS Chem Biol, 2018). Interaction of H2S with metal centers is not only interesting for explaining some of the physiological roles of H2S but also for designing the catalysts for industrial removal of H2S. We also generated a class of iron porphyrins with multiple positively charged substituents which possess excellent catalytic activity for H2S oxidation/removal (WO 2012/175630).
PROTEIN PERSULFIDATION (S-SULFHYDRATION)
We also expanded our interest to newly discovered cysteine modification called protein persulfidation (PSSH). Alongside S-nitrosation (PSNO), S-glutahtionylation (PSSG) and S-sulfenylation (PSOH), persulfidation can represent a new type of redox-switch reaction responsible for the regulation of protein structure and function. However, persulfides are tricky to study as they behave like both thiols and disulfides. This dual nature coupled with the enhanced nucleophilicity (Cuevasanta et al, JBC, 2015) limits their stability in the solution (Yadav et al, JACS, 2015). To selectively label and detect PSSH, we proposed that persulfidation could be selectively detected by the tag switch method (Zhang et al, Angew Chem Int Ed, 2014).
We used this method to study the mechanisms of persulfide formation and their (bio)chemical reactivity. H2S cannot simply react with cysteine residues. This is a redox reaction, so the presence of an oxidant is required. Our work showed that most probable mechanism for PSSH formation would be the reaction of H2S with sulfenic acids, PSOH (Cuevasanta et a, JBC, 2015). The JBC paper was the paper of the week and one of the papers of the year.
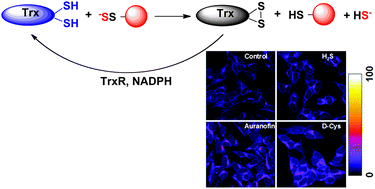
To be able to detect PSSH levels directly in the gel, we advanced the method further by preparing fluorescent cyanoacetate-based probes (Wedmann et al, Chem Sci, 2016). This approach helped us understand how PSSH levels are controlled in the cells. Namely, just as phosphorylation is controlled by dephosphorylation, persulfidation is controlled by depersulfidation. Using our improved tag-switch method we could show that thioredoxin system efficiently cleaves persulfides restoring back the thiols (Wedmann et al, Chem Sci, 2016) in the cells and humans. The method was also successfully used to address the role of protein persulfidation in spinocerebellar ataxia type 3 model in D. melanogaster (Snijder et al, Mol Med, 2015).
In our most recent work (Zivanovic et al, Cell Metab, 2019) we developed a robust method for chemoselective persulfide labelling using dimedone-based probes to show that persulfidation is an evolutionarily-conserved posttranslational modification used by the cells to protect proteins from overoxidation caused by different stressors. Higher persulfidation levels, caused by pharmacological or dietary interventions, lead to better resistance to oxidative stress and longer life.